-
Notifications
You must be signed in to change notification settings - Fork 5
Examples ultraNeuroMorpho2Mesh
ultraNeuroMorpho2Mesh reconstructs high fidelity and geometrically realistic two manifold watertight surface mesh models of neurons from their input morphology skeletons. In this page, we will present a few examples that demonstrate how to use the application.
ultraNeuroMorpho2Mesh creates the same artefacts created by ultraMesh2Mesh; the only difference is the input data source. While ultraMesh2Mesh takes non-watertight input meshes, ultraNeuroMorpho2Mesh takes input neuronal morphologies in .SWC and .H5 file formats. Therefore, we strongly recommend to review the examples of ultraMesh2Mesh to have a complete understanding of how to use the application the best possible way.
This minimal example takes an .SWC neuronal morphology and produces a corresponding watertight mesh.
The following command takes the absolute path of an input morphology, defined by the argument --morphology, and creates an .OBJ mesh (with the same prefix) in the meshes directory that is created in the output directory given by the user by the argument --output-directory.
ultraNeuroMorpho2Mesh --morphology ultraNeuroMorpho2Mesh/input/neuron-1.swc --output-directory ultraNeuroMorpho2Mesh/example-1 --export-obj-mesh --ignore-marching-cubes-mesh --ignore-laplacian-mesh
The input morphology neuron-1.swc is a spatially modified clone of an original morphology of a hippocampal pyramidal neuron that is publicly available on NeuroMorpho.Org (Cell Name: KO-1-DIV-TTb). the morphology editing is performed using the Morphology Editing Toolbox in NeuroMorphoVis. The input morphology is available in the directory examples/ultraNeuroMorpho2Mesh/input/neuron-1.swc. The previous command creates a watertight mesh neuron-1-watertight.obj corresponding to the input morphology to the meshes subdirectory. A side-by-side comparison between the input morphology (left) and the generated mesh (right) is shown in the following image.

-
The intermediate meshes are normally used for comparative analysis and error measurements. It is always advisable NOT to save them to disk using the suppression flags
--ignore-marching-cubes-mesh --ignore-laplacian-mesh. -
The thin branches in the morphology are slightly dilated in the mesh. This is becuase the volume used to voxelize the input morphology has by default a resolution of 512. Increasing the resolution using the argument
--resolution NUMBERor using the--scaled-resolutionflag with 5 microns per voxel--voxels-per-micron 5will eliminate this problem. -
The previous command uses - by default - surface voxelization to only reconstruct the output mesh. Applying the isosurface reconstruction algorithm (either marching cubes or dual marching cubes) to the volume shell created with surface voxelization leads to generate a mesh with multiple partitions. To avoid this problem, it is always advisable to use solid voxelization using the
--solidflag. In the image below, we highlight in a side-by-side comparison the meshes resulting without (light green) and with (light blue) solid voxelization using transparency rendering. -
The resulting mesh is watertight, but it is not optimized. To optimize the mesh, use the
--optimize-meshflag to apply uniform optimization or--optimize-adaptivelyto apply adaptive optimization. -
The soma shape is computed using a physically-plausible approach using the Finite-element method (FEM).


In this example, we address all the notes in the previous one and create an optimized mesh of neuron-2.swc.
ultraNeuroMorpho2Mesh --morphology ultraNeuroMorpho2Mesh/input/neuron-1.swc --output-directory ultraNeuroMorpho2Mesh/example-2 --export-obj-mesh --ignore-marching-cubes-mesh --ignore-laplacian-mesh --ignore-optimized-mesh --project-xy --scaled-resolution --voxels-per-micron 7 --solid --optimize-mesh --optimization-iterations 10 --laplacian-iterations 20


Neurons have different types of neurites or branches: axons, basal and apical dendrites. In certain situations, we would like to highlight a part of the morphology at a specific branching order or completely omit other branches, for example omitting the axons due to their limited significance in reaction-diffusion simulations. We implemented a feature allowing the user to specificy the maximum branching order of a given neuron morphology to mesh.
This is addressed using the following arguments: --axon-branch-order NUMBER , --basal-branch-order NUMBER and --apical-branch-order NUMBER .
The following commands reconstruct multiple neuronal meshes with the respective branching orders 3, 5, 7 and 9.
for i in 3 5 7 9;
do ultraNeuroMorpho2Mesh --morphology ultraNeuroMorpho2Mesh/input/neuron-3.swc --output-directory ultraNeuroMorpho2Mesh/example-3 --export-obj-mesh --ignore-marching-cubes-mesh --ignore-laplacian-mesh --ignore-optimized-mesh --project-xy --scaled-resolution --voxels-per-micron 3 --solid --optimize-mesh --optimization-iterations 10 --laplacian-iterations 15 --basal-branch-order $i --apical-branch-order $i --prefix branch-order_$i; done| # | Maximum Branching Order | Projection |
|---|---|---|
| 1 | 3 |  |
| 2 | 5 |  |
| 3 | 7 |  |
| 4 | 9 |  |
In certain cases, reconstructed morphologies have sampling artifacts, i.e. registering samples with very tiny radii (almost zero). These zero-radius samples can cause severe mesh reconstruction artifacts and can potentially lead to dettaching branches from the final mesh. Therefore, we have added a feature with which users can specify the minimum sample radius in the morphology using the --min-sample-radius NUMBER option.
The following command reconstruct multiple neuronal meshes taking into account different values for the minimum sample radius 0.1, 0.5 and 1.0.
for i in 0.1 0.5 1.0;
do ultraNeuroMorpho2Mesh --morphology ultraNeuroMorpho2Mesh/input/neuron-1.swc --output-directory ultraNeuroMorpho2Mesh/example-4 --export-obj-mesh --ignore-marching-cubes-mesh --ignore-laplacian-mesh --project-xy --min-sample-radius --stats $i; done| # | Minimum Sample Radius | Projection |
|---|---|---|
| 1 | 0.1 microns |  |
| 2 | 0.5 microns |  |
| 3 | 1.0 microns | 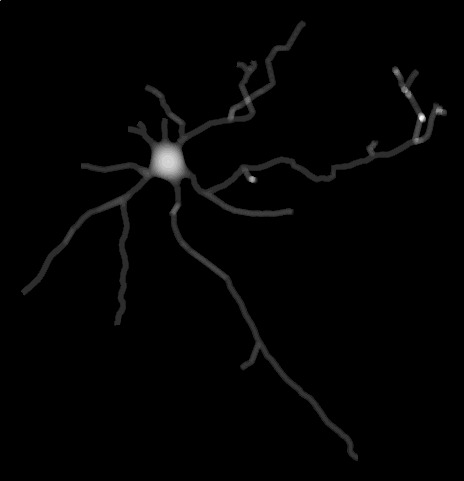 |
-
The default value of the argument
--min-sample-radiusis 0.05 microns. This value has been set based on discussions with the experts. -
The quantitative analysis of the input morphology
neuron-1.swcis shown below. The minimum sample radius is verified to be 0.07 microns, while the maximum sample radius is 0.43 microns. Using values greater than the maximum sample radius like 0.5 and 1.0 microns is only for demonstrative purposes. But in reality, this value must be set very carefully to avoid any misleading results if the generated meshes are used in reaction-diffusion simulations.
Morphology Stats. [ neuron-1 ]
* Bounding Box: | [154.72287, 160.52518, 58.35493]
* pMin: | [-100.22283, -109.95874, -22.06759]
* pMax: | [54.50004, 50.56643, 36.28735]
* Number Samples | 371
* Number Sections | 32
* Number Segments | 365
* Morphology Length | 724.551453
* Surface Area | 1979.219849²
* Volume | 644.346985³
* Distributions:
* Samples Radii | [Min.: 0.07000, Max.: 1.87500, Avg.: 0.43155]
* Segments Lengths | [Min.: 0.14000, Max.: 11.86742, Avg.: 2.34398]
* Sections Lengths | [Min.: 0.38897, Max.: 80.50992, Avg.: 22.64223]
* Segments Surf. Areas | [Min.: 1.90918, Max.: 18.95555, Avg.: 9.96011]
* Sections Surf. Areas | [Min.: 2.31174, Max.: 218.69467, Avg.: 61.85062]
* Segments Volumes | [Min.: 0.15705, Max.: 10.69837, Avg.: 3.41087]
* Sections Volumes | [Min.: 0.14950, Max.: 90.11069, Avg.: 20.13584]